Research summary
Research outline
“Why not watching directly?” We have proven that individual fluorescently-labeled proteins can be visualized in living cells. Using this technique, we have elucidated dynamic coupling between cytoskeletal remodeling, cell signaling and mechanical forces. For example, mDia1 and other members of formin homology proteins rapidly regenerate actin filaments in mechanically perturbed cells. Formins exert torque on their polymerizing actin filament and protect it from the depolymerizing factor cofilin. Formins thus function as cellular anti-mechanostress machineries. At the leading edge of the cell, some newly-assembled actin filaments rapidly disassemble by a mechanism distinct from widely-accepted ‘treadmilling’. These cell edge-contacting F-actin functions as a ‘Brownian ratchet force sensor’ to direct cell protrusion toward traction force. We also apply high resolution imaging to elucidate the mechanism of action of target-based drugs. Therapeutic tyrosine kinase inhibitors allosterically perturb autoinhibition in Src-related kinases, which may lead to unwanted paradoxical growth of cancer cells by drug treatment. All of these examples of our discoveries might have been overlooked by conventional ‘phenotype-based analysis’ commonly employed in biological and medical researches.
Cells undergo dynamic shape change during development, immune response, cancer invasion, neural network formation and so on. Because such dynamic processes operate in a complex manner, it has been difficult to precisely dissect the role of individual molecules just by observing phenotypes. Our direct real-time viewing of ‘live’ molecular kinetics has brought a powerful tool to elucidate how the complex system of life operates in live cells and tissues. Our laboratory also invented ultra-multiplexed, high-density labeling super-resolution microscopy IRIS and extends its applications to the nervous system and animal disease models. We are elucidating ‘real time and space’ of signal- and mechano-transducing molecules in the hope of developing new strategies for tissue engineering and drug seed discovery.
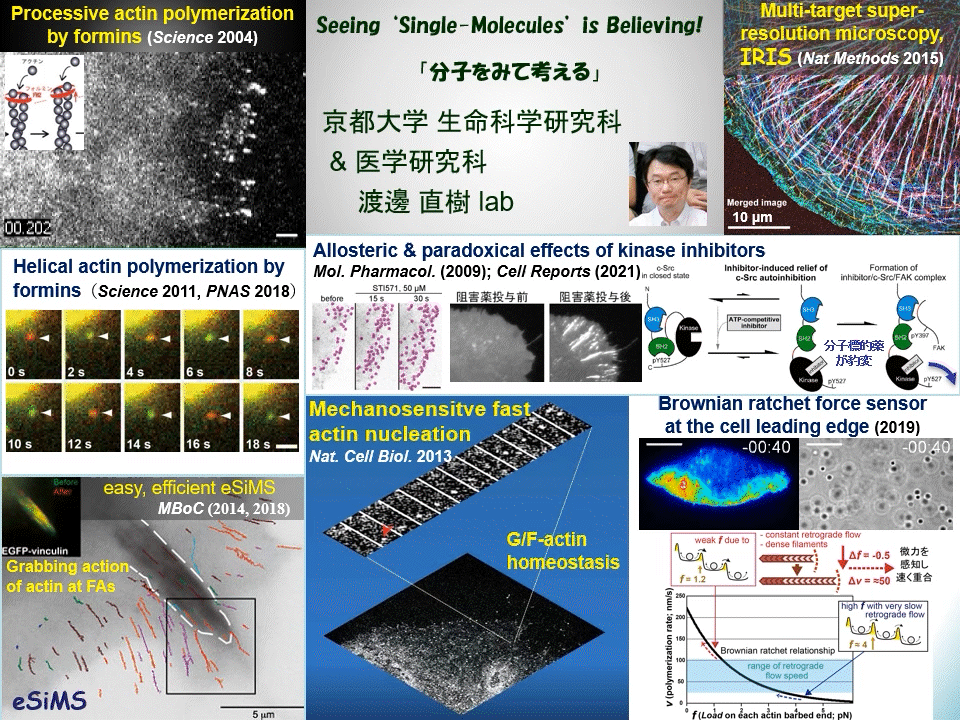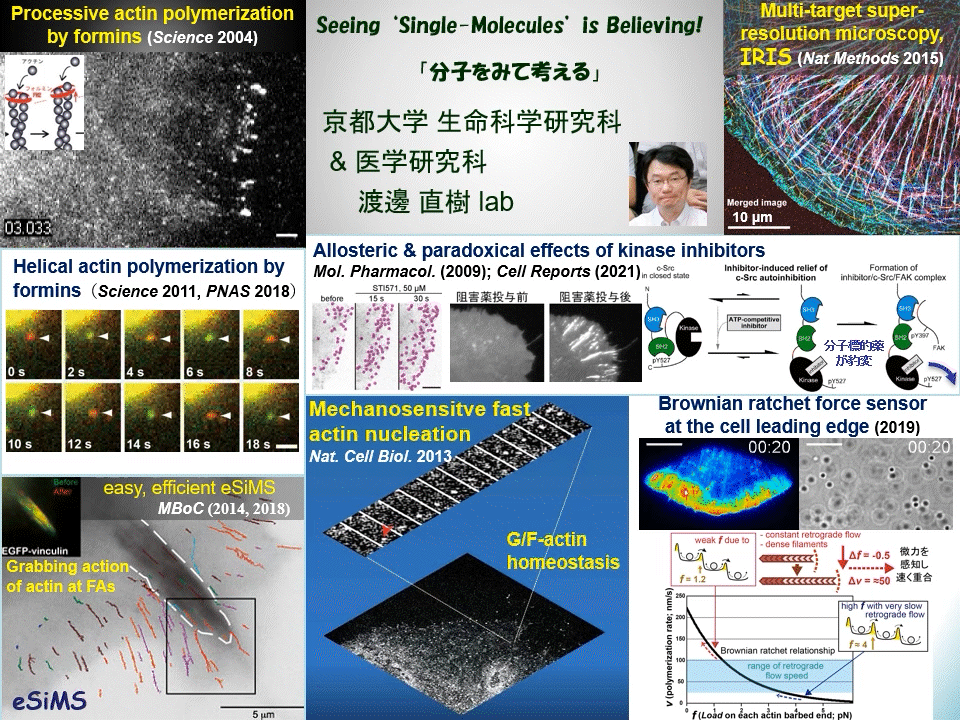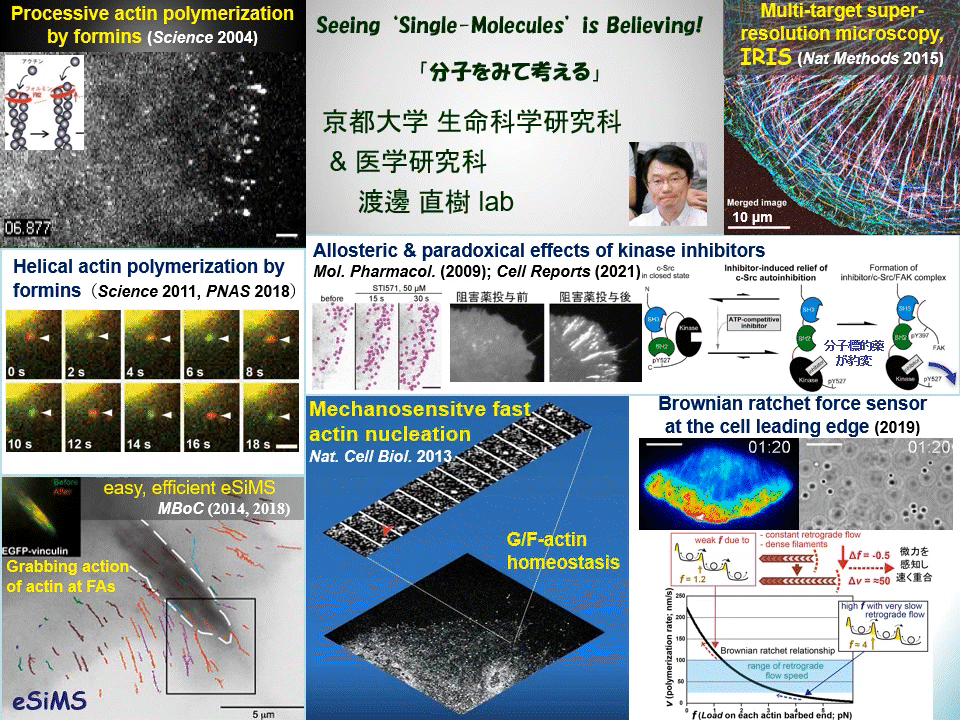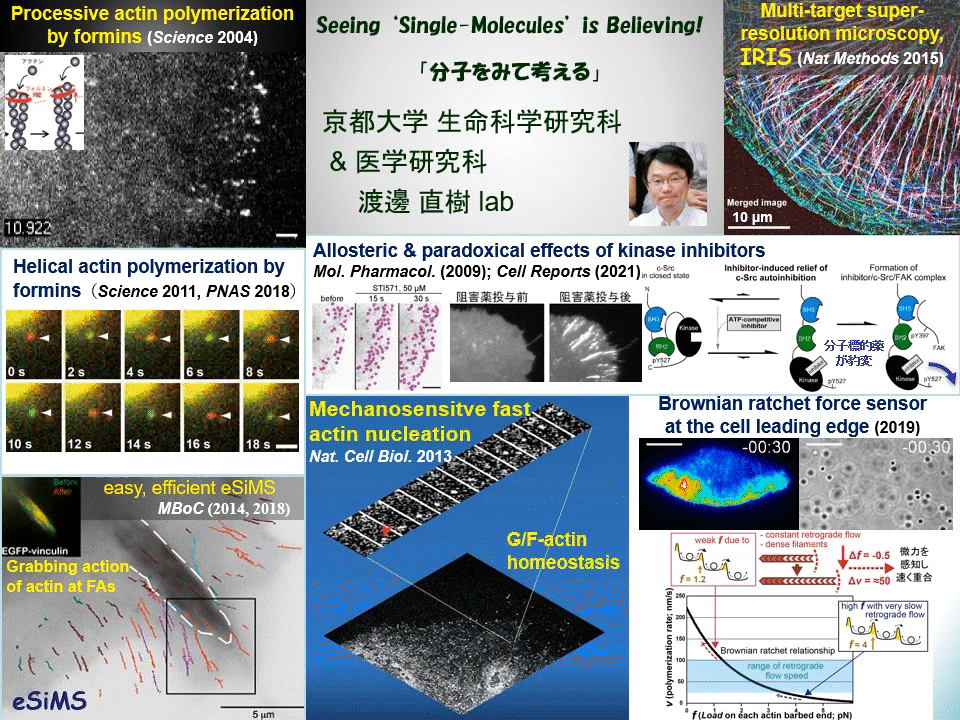
Main themes
- Single-molecule imaging of mechanotransduction and its underlying signaling
- Elucidation of tissue remodeling process by multiplexed super-resolution microscopy IRIS
- Imaging-based elucidation of the mechanism of action of drugs for drug discovery
Member
|
WATANABE, NaokiProfessor |
watanabe.naoki.4v*kyoto-u.ac.jp See faculty information |
|---|---|
|
YAMASHIRO, SawakoAssociate Professor |
yamashiro.sawako.5c*kyoto-u.ac.jp See faculty information |
|
MIYAMOTO, AkitoshiAssistant Professor |
miyamoto.akitoshi.2c*kyoto-u.ac.jp See faculty information |
- Please note that the @ symbol has been replaced by *.
Access
Faculty of Medicine Campus, Faculty of Medicine Bldg. A



