Member
|
KITAJIMA, TomoyaVisiting Professor |
tomoya.kitajima*riken.jp See faculty information |
|---|---|
|
FUJISAWA, ShigeyoshiVisiting Professor |
shigeyoshi.fujisawa*riken.jp |
|
TAKASATO, MinoruVisiting Associate Professor |
minoru.takasato*riken.jp See faculty information |
|
OBATA, FumiakiVisiting Associate Professor |
fumiaki.obata*riken.jp See faculty information |
|
KONAGAYA, YumiVisiting Associate Professor |
yumi.konagaya*riken.jp See faculty information |
|
KONDO, TakefumiVisiting Associate Professor |
takefumi.kondo*riken.jp See faculty information |
|
TAKEOKA, AyaVisiting Associate Professor |
aya.takeoka*riken.jp |
- Please note that the @ symbol has been replaced by *.
Access
Kitajima lab
RIKEN(Kobe Campus) Center for Biosystems Dynamics Research, Developmental Biology Building C
Fujisawa lab
RIKEN Center for Brain Science(Wako Campus), CBS Neural Circuit Genetics Research Building
Takasato lab
RIKEN(Kobe Campus) Center for Biosystems Dynamics Research, Developmental Biology Building A
Obata lab
RIKEN(Kobe Campus) Center for Biosystems Dynamics Research, Developmental Biology Building A
Konagaya lab
RIKEN(Kobe Campus) Center for Biosystems Dynamics Research, Developmental Biology Building A
Kondo lab
RIKEN(Kobe Campus) Center for Biosystems Dynamics Research, Developmental Biology Building C
Takeoka lab
RIKEN Center for Brain Science(Wako Campus), CBS Neural Circuit Genetics Research Building
Kobe Campus Map
Kitajima lab research summary
Research outline
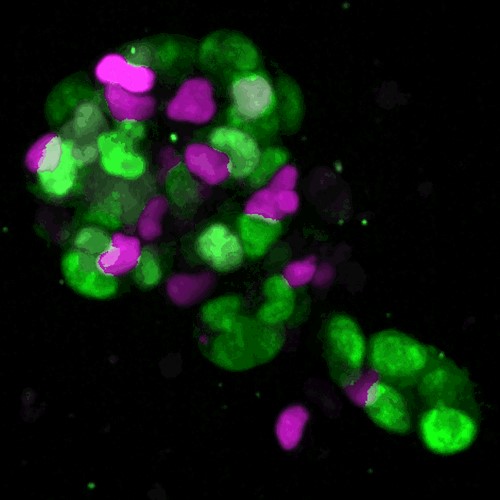
The oocyte becomes an egg through meiosis. The egg fertilizes with a sperm and undergoes repeated cell divisions to give rise to an entire body. We study chromosome segregation during meiosis in oocytes and during mitosis in fertilized eggs, taking advantage of techniques for high-throughput and high-resolution live imaging of mouse oocytes combined with micromanipulation and genetic engineering methods. The first cell division that oocytes undergo is meiosis I. Chromosome segregation in this division is error-prone and the rate of errors increases with maternal age. Subsequently, chromosomes are segregated in meiosis II upon fertilization, and then segregated again in mitosis after DNA replication. We will reveal distinct mechanisms for chromosome segregation during these subsequent but fundamentally different cell divisions. By uncovering the mechanism of chromosome segregation during meiosis I in oocytes, we understand why oocyte meiosis I is error-prone and related to age. Comparing the mechanisms in meiosis I with those found in meiosis II and mitosis may provide insights into the capacity of cells to flexibly use different strategies for chromosome segregation. The findings will be exploited to collaborative studies with reproductive medicine.
Main themes
- Molecular mechanisms of meiotic chromosome segregation in mouse oocytes
- Live imaging of chromosome dynamics in oocytes
- Aging-associated deterioration of chromosome segregation in oocytes
Fujisawa lab research summary
Research outline
Main themes
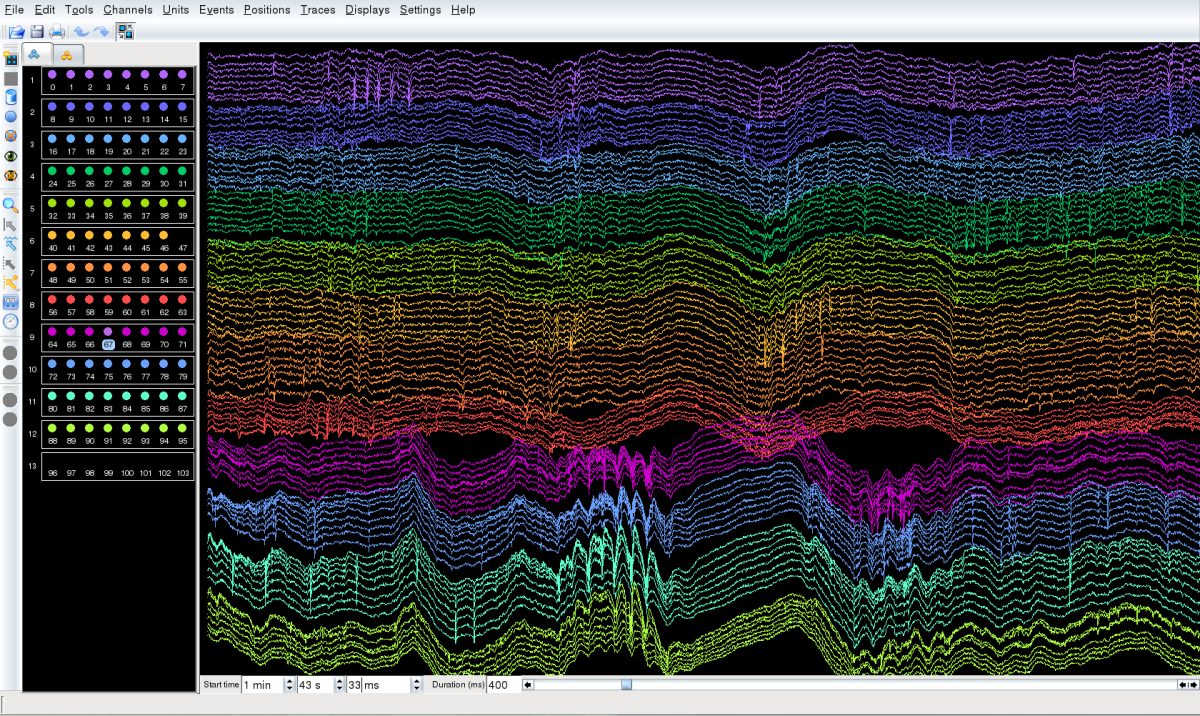
- To elucidate the mechanism of spatial navigation in the hippocampus and entorhinal cortex.
- To clarify the role of oscillations in the information processing of neuronal circuits.
- Development of technology for manipulating neuronal activity by combining optogenetics and electrophysiology
Takasato lab research summary
Research outline
What do you think is the ultimate goal of regenerative research using human pluripotent stem cells? We think that is to recreate a whole replaceable organ in vitro via directed differentiation. In the previous study, we developed a protocol by which human pluripotent stem cells can be differentiated into the intermediate mesoderm that self-organizes kidney organoids. Kidney organoids comprises all anticipated renal tissues, including nephrons, collecting duct, blood vessels and renal interstitium, although, they are still far from the real human kidney in their size, tissue complexity, maturity and functionality. By precisely recapitulating the developmental process of human kidney in directed differentiation of human pluripotent stem cells, we are trying to achieve the ultimate goal, generation of a three-dimensional urinary tract including the kidney and the bladder that is functional and transplantable to the patients. We also study about how to recapitulate the development of other organs that connect to the kidney. We appreciate knowledge from basic developmental biology that is essential for such regenerative studies; therefore, we are highly interested in studies of human embryology. Utilizing our unique technology that generates hPSCs-derived kidney organoids from the pluripotent stage in vitro, we are especially investigating developmental mechanisms of human mesoderm and kidney.
Main themes
- Elucidate the mechanisms of cell fate determination of pluripotent stem cells using single-cell RNA-seq analysis.
- Interact epithelial and mesenchymal stem cells to form organs by self-organization.
- Direct the differentiation of human ES/iPS cells to generate 3D tissues so called “organoids” such as kidney and bladder
- Generate kidney organoids from human iPS cells which were derived from patients with hereditary kidney disease and establish disease models.
- Develop methods to promote vascularization and maturation of organoids to enable their application usages.
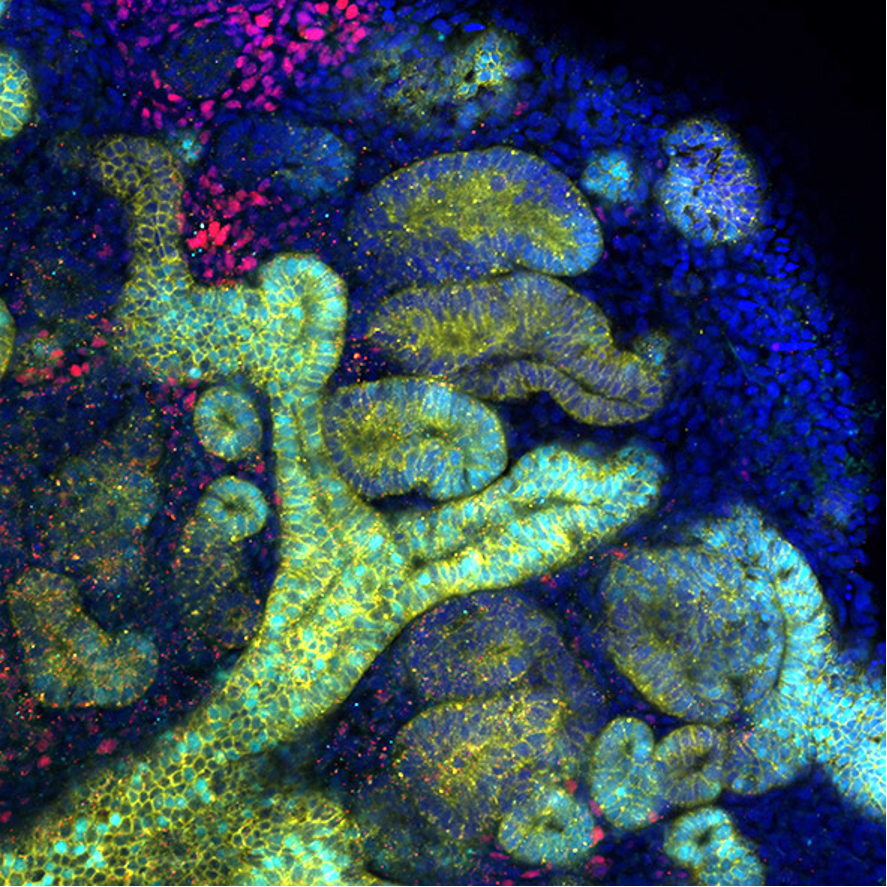
Kidney organoids derived from human iPS cells: The organoid contains two kidney progenitors, the ureteric tree (yellow with cyan) and nephron progenitor (red), as well as developing nephrons (yellow).
Movie of research introduction
Confocal microscopic Z-stack images from the bottom to the top of kidney organoids. Developing nephrons are segmented into 4 compartments, including the collecting duct (yellow and green), distal tubule (yellow only), proximal tubule (red) and glomerulus (green only).
Obata lab research summary
Research outline
All animals require constant nutrition from their diet as they grow and age. Our healthspan is greatly influenced by the dietary environment. Diet contributes to the metabolic and physiological homeostasis of animals directly as nutrients or indirectly via gut bacteria, but our understanding of the detailed molecular mechanisms is limited. Our laboratory studies the physiological functions of various nutrients and gut bacteria. We are also trying to elucidate the mechanisms by which transient dietary intake during development influences health status throughout life. We use the short-lived fruit fly Drosophila melanogaster to elucidate the universal “dietological” mechanisms that regulate the ageing and lifespan of organisms.
Our research focuses on lifespan as an indicator of an individual’s overall capacity for homeostasis. We need to carefully unravel the mechanisms of how and which factors determine lifespan. In conducting research on ageing, it is reasonable to use simpler organisms with shorter life spans. We use cheap, handy, and a sophisticated genetic model Drosophila, which has a lifespan of only two months. It has few ethical problems and is ideal for life-span measurements, which requires a huge number of animals. Therefore, Drosophila as a simple model organism, is the best tool to understand the universal and fundamental mechanisms of the organism.
Until now, biomedical studies seem to have aimed at how to cure and/or avoid disease. However, being healthy and not being sick are similar but different. It seems to us that living organisms inherently have the power to try to become healthy. In our research, we are trying to clarify the basic mechanisms of homeostasis.
Main themes
- Elucidation of functions of each nutrient and gut bacterial species
- Elucidation of the mechanism by which early life environment alters homeostasis
- Elucidation of the amino acid regulation of healthspan
- Molecular basis of thermotolerance
- Molecular basis of feeding/blood feeding
Konagaya lab research summary
Research outline
Adult tissues are constantly undergoing turnover to maintain tissue homeostasis. The intestinal epithelium is one of the most rapidly renewing tissues, with new epithelial cells replacing old ones every 3 to 5 days in mice. Precise control of cell proliferation during intestinal epithelial differentiation is essential for maintaining tissue homeostasis, and its breakdown can lead to tissue atrophy or cancer.
In the intestinal epithelium, Lgr5-positive stem cells divide every 24 hours to produce transit-amplifying cells. These transit-amplifying cells then divide 4 to 6 times at a rate approximately twice as fast as Lgr5-positive stem cells. After that, the transit-amplifying cells slow down the cell cycle as they terminally differentiate into absorptive or secretory cells, and eventually exit the cell cycle. Remarkably, despite descriptions of the cell cycle in the intestinal epithelium as mentioned above being reported more than 25 years ago, the elucidation of the molecular mechanisms by which intestinal epithelial cells regulate cell proliferation rates during differentiation has made little progress, likely due to technical constraints.
Therefore, our laboratory uses mouse intestinal epithelium as a model to clarify the molecular mechanisms that regulate cell proliferation rates as stem cells transit through transit-amplifying cells to terminally differentiated cells. In particular, we use multiplexed imaging of intestinal organoids and tissues, quantitative analysis, and mathematical modeling. These studies will reveal universal and fundamental therapeutic targets for tissue homeostasis disorders.
Main themes
- Investigation of the coordinated control of cell proliferation and differentiation in mouse intestinal epithelium
- Analysis of the role of mechanical sensing in stem cell maintenance in mouse intestinal epithelium
- Development of fluorescent reporters using machine learning algorithms for protein structure prediction
Kondo lab research summary
Research outline
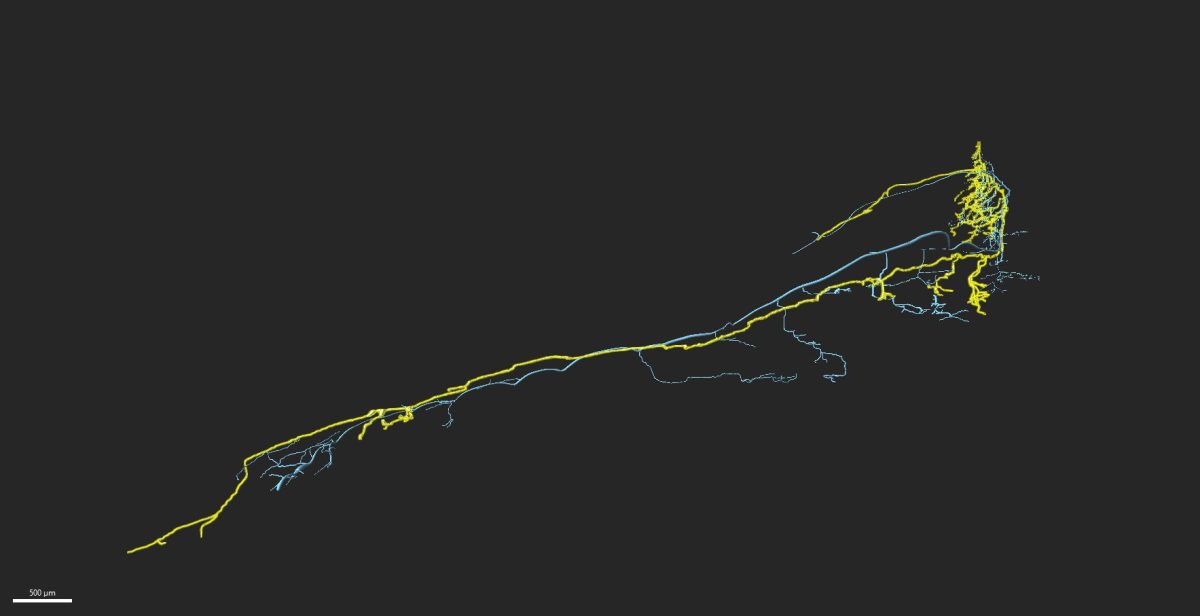
Embryonic development is a dynamic and beautiful phenomenon, progressing with remarkable accuracy. However, some degree of uncertainty is observed at more detailed levels, such as the cellular and gene levels. We aim to unravel the principles that ensure the accuracy of an overall developmental process.
An animal’s body comprises the combination of organs acquiring appropriate ‘form’ and ‘function.’ During animal development, epithelial tissues, which consist of a single layer of cell sheets, repeatedly grow and deform to shape an appropriate form. The function of an organ depends on the properties of cells, and cells appropriately differentiate to fulfill the physiological roles of their constituent organs. In the developmental process, these physical and biochemical processes proceed simultaneously. Therefore, to understand the self-organization mechanism in organogenesis, we believe that it is crucial to comprehend the dynamics of each process, how they mutually influence each other in time and space, and how this crosstalk achieves the harmony of form and function. We consider development as a multi-hierarchical network system of genome-cell-tissue, based on bidirectional information feedback between gene expression, cell differentiation, and morphogenesis.
Our laboratory aims to reveal the diversity and universality of this feedback mechanism across the genome-cell-tissue hierarchy by quantitatively analyzing the dynamics at each level utilizing techniques such as single-cell genomics and imaging and large-scale data analysis.
Main themes
- Elucidation of the rules governing how cells process the gene expression information to drive morphogenesis.
- Understanding the mechanism by which tissue morphological changes control gene expression and cell differentiation.
- Comprehensive analysis of cell differentiation dynamics in entire embryonic development.
- Development and application of single-cell and spatial genomics analysis techniques.
Takeoka lab research summary
Research outline
Main themes
- Spinal learning and memory
- Sensory integration for motor adaptation
- Sensorimotor circuit organization and function